A Spatial Neighborhood Methodology for Computing and Analyzing Lymph Node Carcinoma Similarity in Precision Medicine
March 1st, 2020
Categories: Applications, Software, Visualization, Visual Analytics, Visual Informatics, Data Science
Authors
Luciani, T., Wentzel,A., Elgohari, B., Elhalawani, H., Mohamed, A.S., Canahuate, G., Vock, D.M., Fuller, C.D., Marai, G.E.About
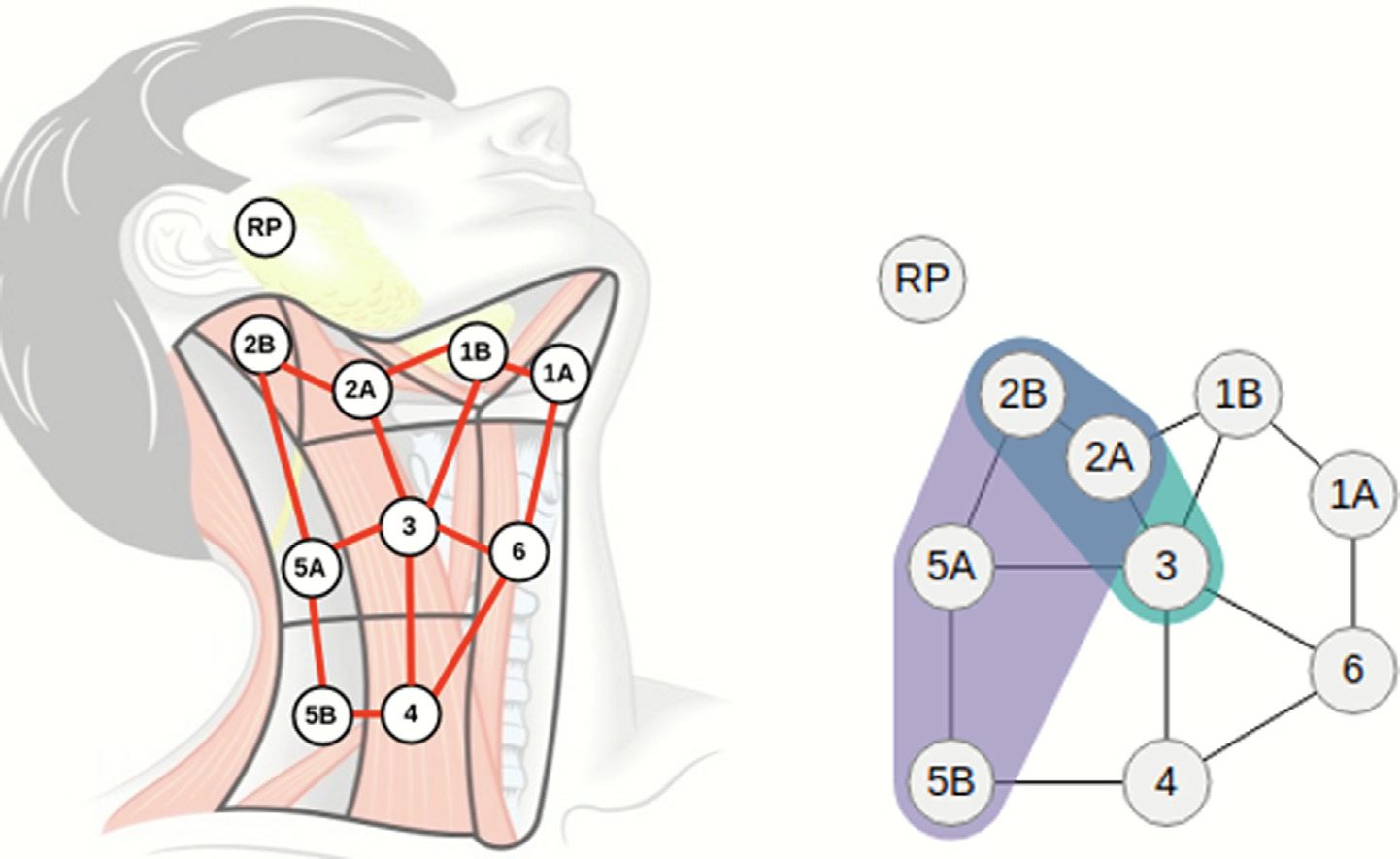
Precision medicine seeks to tailor therapy to the individual patient, based on statistical correlates from patients who are similar to the one under consideration. These correlates can and should go beyond genetics, and in general, beyond tabular or array data that can be easily represented computationally and compared. For example, in many types of cancer, cancer treatment and toxicity depend in large measure on the spatial disease spread - e.g., metastasizes to regional lymph nodes in head and neck cancer. However, there is currently a lack of methodology for integrating spatial information when considering patient similarity. We present a novel modeling methodology for the comparison of cancer patients within a cohort, based on the spatial spread of the lymph nodes affected in each patient. The method uses a topological map, bigrams, and hierarchical clustering to group patients based on their similarity. We compare this approach against a nonspatial (categorical) similarity approach where patients are binned solely by their affected nodes. We present similarity results on a 582 head and neck cancer patient cohort, along with two visual abstractions for analysis of the results, and we present clinician feedback. Our novel methodology partitions a patient cohort into clinically meaningful groups more susceptible to treatment side-effects. Such spatially-aware similarity approaches can help maximize the effectiveness of each patient’s treatment.
Funding: NIH NCI-R01CA214825, NIH NCI-R01CA2251, and NIH NLM-R01LM012527
Resources
URL
Citation
Luciani, T., Wentzel,A., Elgohari, B., Elhalawani, H., Mohamed, A.S., Canahuate, G., Vock, D.M., Fuller, C.D., Marai, G.E., A Spatial Neighborhood Methodology for Computing and Analyzing Lymph Node Carcinoma Similarity in Precision Medicine, Journal of Biomedical Informatics, vol 5, March 1st, 2020. https://www.sciencedirect.com/science/article/pii/S2590177X20300019